Our initial investigations will focus on heart failure and will explore the paradigm that disease pathogenesis is governed by a multi-hit model that can be intervened upon through individualized precision medicine strategies. We postulate that patient specific genomic variants govern disease susceptibility and initiation (primary hit), and that maladaptive inflammatory responses to tissue injury and/or dysfunction (secondary hits) contribute to disease progression. Furthermore, we believe that the development of therapeutics that target each of these mechanisms will revolutionize the care of heart failure patients.
Functional Genomics of Dilated Cardiomyopathy (DCM): Single Cell Mapping of the Human Heart
DCM represents a prominent cause of heart failure in children and adults and is the most common diagnosis in patients listed for heart transplantation. Mutations are increasingly being identified in patients with DCM, a disease previously thought to be idiopathic. Little is known about whether patients who carry mutations in different genes for DCM display distinct disease phenotypes and/or different clinical outcomes including responses to medical therapies.
We believe that DCM should be reclassified based on mutations that give rise to phenotypes. If genetic subtypes of heart failure can be identified based on the mutations found in DCM patients, then unique therapeutic interventions may be designed and used to treat DCM.
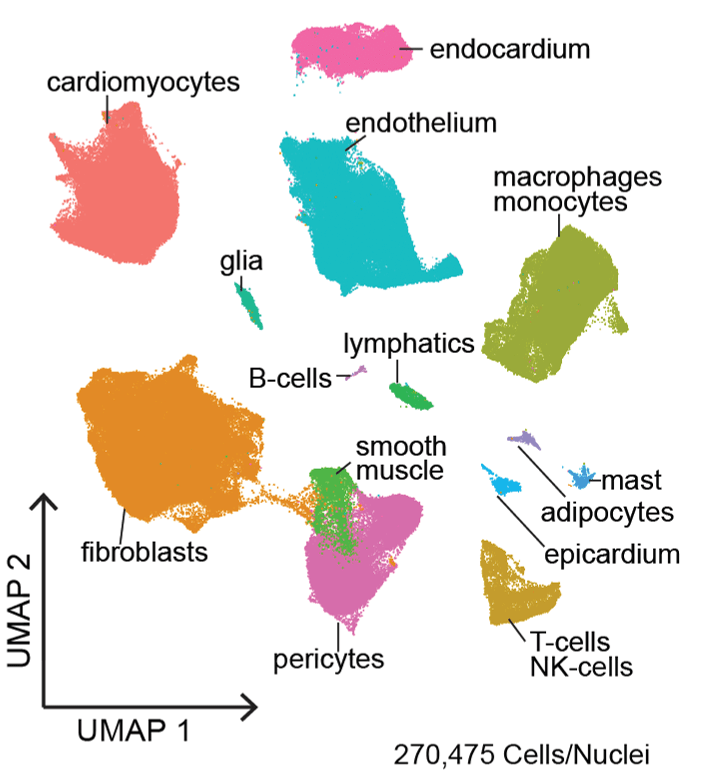
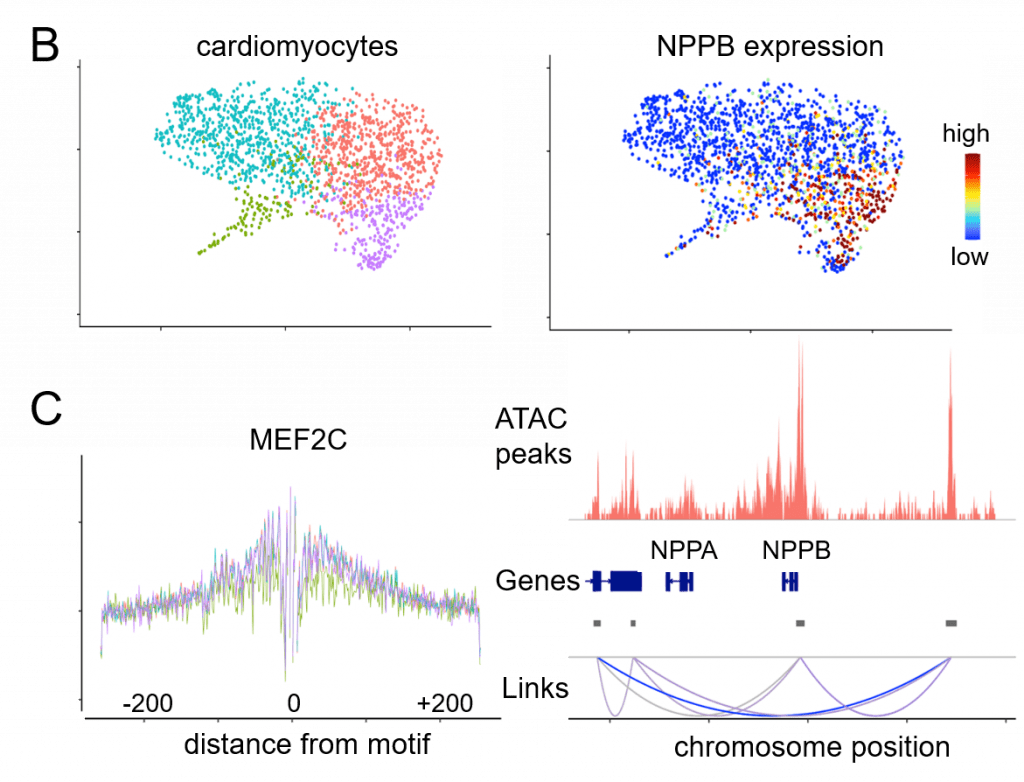
The Translational Cardiovascular Biobank and Repository (TCBR) and Washington University Inpatient Heart Failure Registry represent two clinical resources that have been developed to address the genetic subsets of heart failure though phenotypic and pathological analyses. Single cell and single nucleus RNA and ATAC sequencing, spatial transcriptomics, proteomics, and lipidomics are innovative technologies used within the laboratories to understand specific features, underlying gene regulatory networks, and effector mechanisms of cell types that emerge during heart failure across different etiologies.
Novel approaches utilizing explanted tissues, engineered heart tissues and new mouse models provide exciting opportunities to obtain new and unprecedented information into how individual DCM mutations influence mechanical, structural, and electrophysiological properties that contribute to heart failure pathogenesis.
Precision Therapeutics for Genetic Cardiomyopathies
The ultimate goal of CPRi is to develop new diagnostics and therapeutics for patients. Given that the majority of DCM patients harbor heterozygous missense mutations that display reduced or absent activity, we are identifying compounds that selectively bind to mutant DCM proteins. We use DNA encoded libraries that contain 1000x more entities than traditions small molecule libraries. This technology binds the compounds within the library by affinity allowing to sequence the codes and identify compounds that bind that DCM mutant.
Troponin mutant complexes are used to establish the process, but many DCM variants can be used. The compounds that selectively bind to the DCM mutant proteins ideally will lead to the development of new therapeutics that precisely target the genetic underpinnings of DCM. We have leveraged this technology to identify a new thin filament activator.
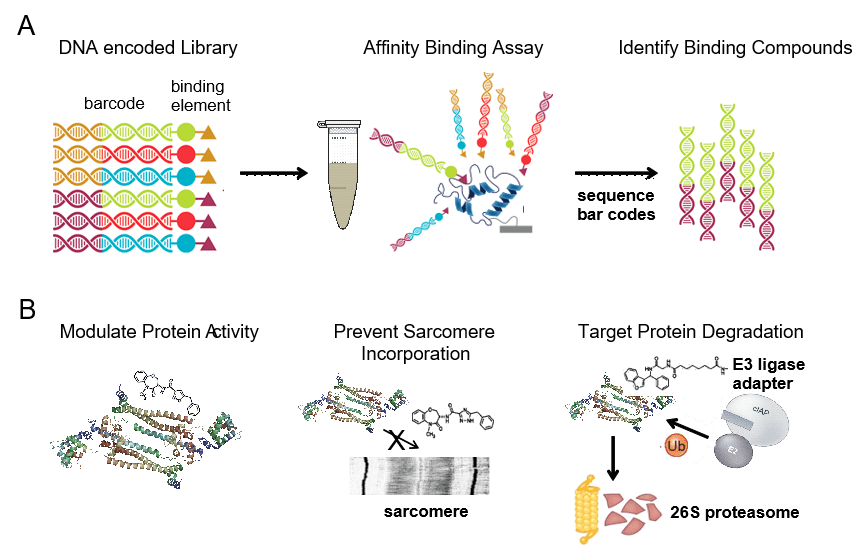
Targeting inflammation
Inflammation is considered an important mechanism that contributes to both nonischemic and ischemic cardiomyopathies. In recent years, a renewed interest in targeting the immune system in cardiovascular disease has emerged. This resurgence is powered by the discovery of the cellular mediators of inflammation in the heart and the identification of the mechanisms that orchestrate their activation and damaging effector functions.
We have established that a specific subset of macrophages (identified based on the expression of C-C Chemokine Receptor 2, CCR2) are key mediators of cardiac inflammation and heart failure progression. From a precision medicine perspective, we have defined targetable signaling events that govern the activation of CCR2+ macrophages and successfully designed a molecular imaging probe (compatible with PET/CT or PET/MRI) to visualize CCR2+ macrophages in patients to facilitate the identification of individuals most likely to benefit from therapeutics targeting this cell population.
Using genetic loss of function models in mice, we have established that CCR2+ macrophages are activated through concomitant engagement of multiple innate immune receptors including Toll-like receptors (TLRs) and the Interleukin 1 receptor (IL1R) that converge on a signal transduction pathway primarily transduced through MYD88 and NF-kb signaling. We are developing targeted therapeutics that prevent the activation and effectors of CCR2+ macrophages. These agents will be tested across cardiovascular conditions that are driven by this inflammatory axis.
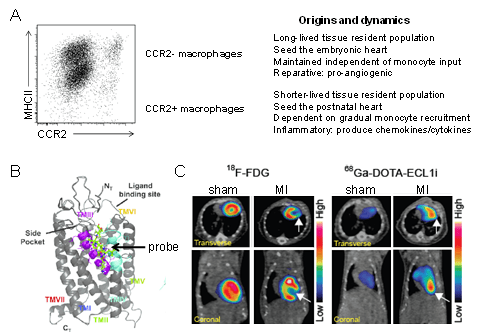
Figure 4. Cardiac macrophage diversity. A ,Flow cytometry plot and schematic depicting the origins, dynamics, and functions of macrophage populations within the heart. B, Model showing that the ECL1i peptide imaging probe binds to CCR2 in an allosteric position. C, PET/CT images demonstrating robust ECL1i tracer uptake within the infarct 4 days following ischemia reperfusion injury. FDG imaging identifies the infarct region.
Elucidating the Cellular and Transcriptional Landscape of Myocarditis and Other Cardiovascular Pathologies
We are deploying an arsenal of molecular pathology tools to dissect the pathogenesis of cardiovascular diseases that lack high fidelity experimental models. These include giant cell myocarditis, cardiac sarcoidosis, cardiac amyloidosis, advanced coronary artery disease, and many others. The ultimately goal is to build testable hypotheses and identify targetable mechanisms based on analysis of patient specimens.